The seed region is located in the center of the crRNAtarget duplex, where it is more sensitive to mismatches than the nonseed region (Abudayyeh et al, 16) Single mismatch can be fully tolerated by C2c2, but if double mismatches are located in the seed region, C2c2 is unable to cleave the singlestrand RNA;This is consistent with the observation that the mismatch of the twocrRNA nucleotide adjacent to PAM is intolerable while offtarget DSB could occur in the DNA sequence homology to the seed region Figure 2 PAMbinding of Cas9/gRNA complex and initiation of crRNA/target DNA hybridizationLength of the seed region The seed region is located in the center of the crRNAtarget duplex, where is more sensitive to mismatches than the nonseed region Its length should not be greater than the length of the target complementarity region of crRNA

Synthetic Crispr Rna Cas9 Guided Genome Editing In Human Cells Pnas
Seed region of a crrna
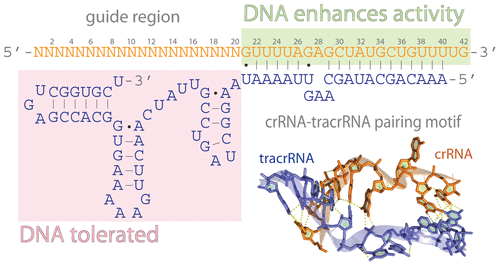
Seed region of a crrna-Interestingly, the 3′ end of the ordered region of crRNAspacer, nt 11–13, which forms part of the central "seed" region previously identified, exits the NUC lobe forming an Aform conformation that appears more solvent accessible than the preceding 10 nt of the crRNAspacerOnce the Cas9gRNA complex binds a putative DNA target, the seed sequence (810 bases at the 3′ end of the gRNA targeting sequence) will begin to anneal to the target DNA If the seed and target DNA sequences match, the gRNA will continue to anneal to the target DNA in a 3′ to 5′ direction



The Influence Of Mismatches Between The Crrna And Target Dna On Binding Download Scientific Diagram
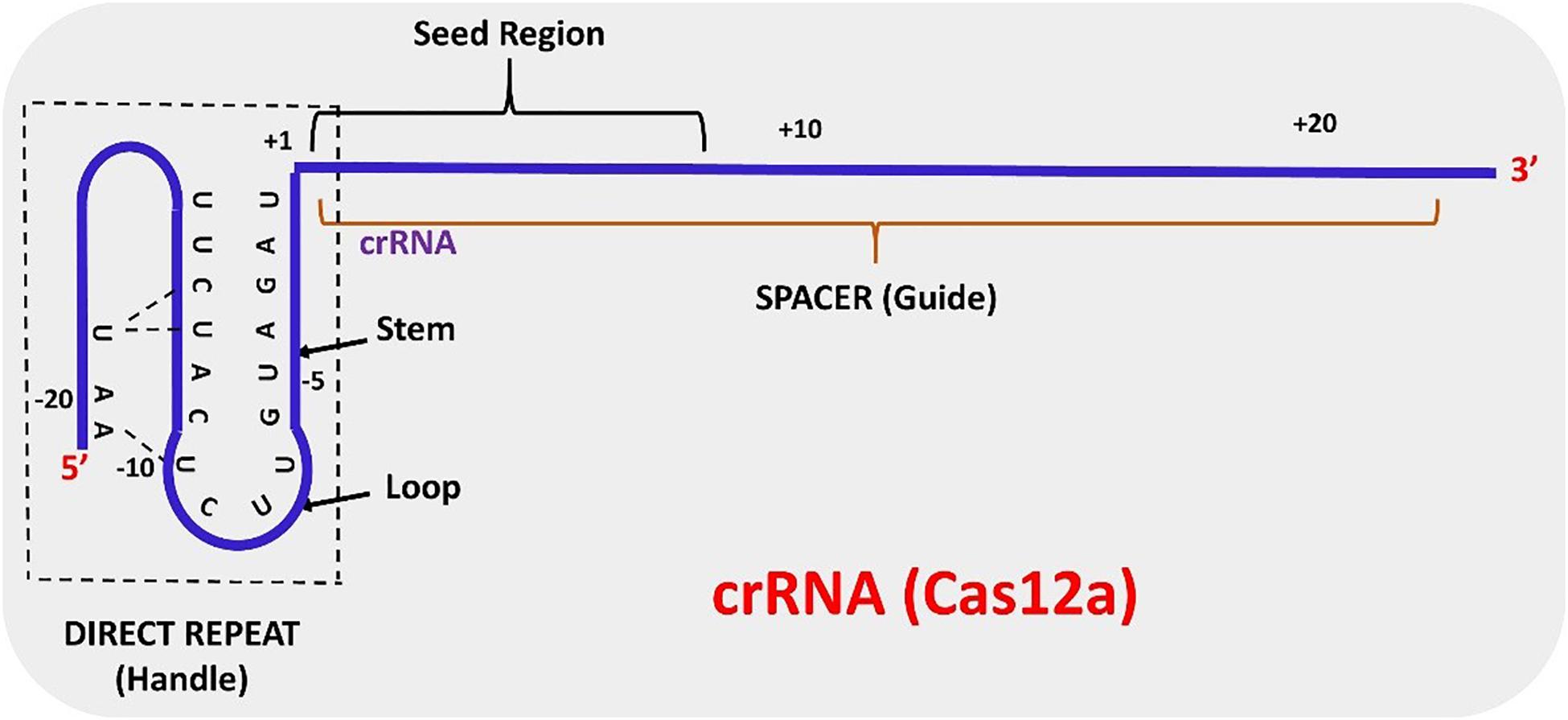
(v) The complex highlevel structure that may decrease the available of target region should be avoidedSeed region flanks at the initial part of the spacer and plays a notable role in the target specificity of CRISPR–Cpf1 system (Fig 3) Fig 3 Schematic representation of a mature Cpf1 crRNAC2c2 can even tolerate three consecutive mismatches in the nonseed region ( Abudayyeh et al , 16 )
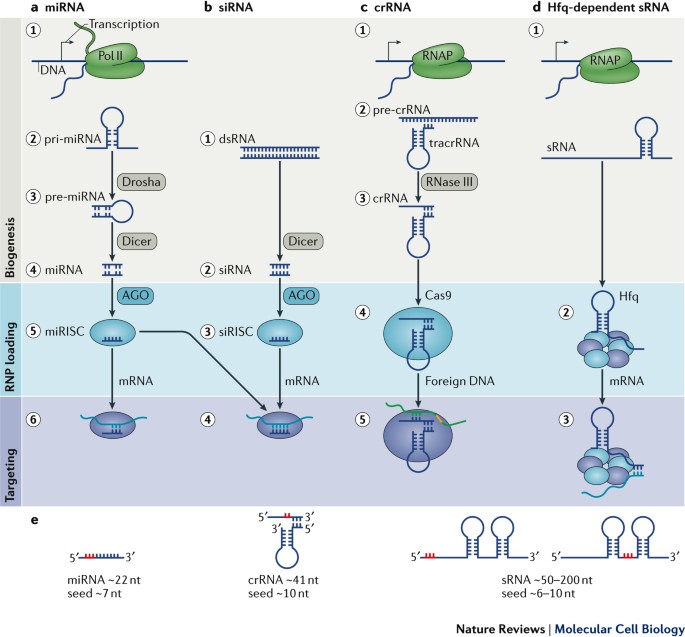
Binding of central seed region in gRNA crucial for Cas13 mediated cleavage (A) Schematic of central seed region of the guide in the crRNA binding the protospacer (target) region in the RNABution of mismatches between crRNA and activatorRNA affect LbuCas13a binding affinity and HEPNnuclease activity Mismatches in the middle seed region (positions 9–12) of the crRNA spacer result in the largest defect in binding affinity In contrast, mismatches elsewhere in the crRNA result in only small reductions of binding affinitySequence recognition and structure of synthetic crRNA and tracrRNA A The target recognition part of the crRNA consists of nt, whereas the seed region is the 10 nt initiating the DNA binding nucleation B The tracrRNA consists of 67 nt comprising an antirepeat region complementary to the crRNArepeat and stem loops responsible for Cas9 binding
In type II CRISPR systems, the seed region has been defined as the PAMproximal 10–12 nucleotides located in the 3′ end of the nt spacer sequence (15, 47, 48, 94) Mismatches in this seed region severely impair or completely abrogate target DNA binding and cleavage, whereas close homology in the seed region often leads to offtarget binding events even with many mismatches elsewhere ( 78 )CrRNA and target RNA can affect Cas13a transcleavage activity in a number and positiondependent manner, especially within the seed region (9~14 nt);TargetcrRNA mismatches have different effects on antiplasmid and antiphage immunity Type I and II CRISPRCas immunity requires the presence of a "seed" region within the target sequence Complementarity between the seed and the spacer sequence in the crRNA guide is essential for immunity ( 5, 10, 11, 13, 14 )



One Or Two Mismatches In The Seed Can Be Tolerated For Crispr Activity Download Scientific Diagram
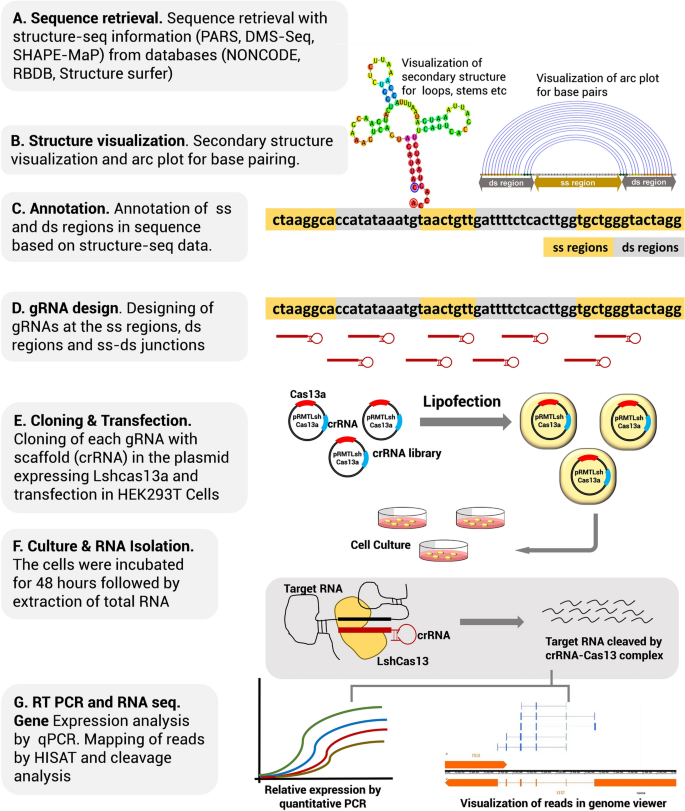


Structure Based Design Of Grna For Cas13 Scientific Reports
Seed sequence The seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the " seed sequence " has to be perfectly complementaryIn particular the seed region (ie, crE2M, crE2N, and crE2R) Minimal RNA content was achieved with as few as six RNA residues in the seed (crE2U and crE2V), although activity was reducedThe structure shown (Top) forms when crRNAguided Cascade complex recognizes doublestranded DNA containing the protospacer (B) The sequence of the g8 protospacer, PAM, and the seed region is shown (Top) Below, point mutations selected as spontaneous escape phages on lawns of cells expressing g8 crRNA and also engineered by sitespecific



Determining The Specificity Of Cascade Binding Interference And Primed Adaptation In Vivo In The Escherichia Coli Type I E Crispr Cas System Mbio



Determining The Specificity Of Cascade Binding Interference And Primed Adaptation In Vivo In The Escherichia Coli Type I E Crispr Cas System Abstract Europe Pmc
Although mismatches between the crRNA and the target DNA are allowed at some positions, the 3' end of the protospacer needs to be fully complementary at the socalled seed region, corresponding to positions 1 to 5 and positions 7 to 8 of the protospacer3 Third, the target DNA needs to be negatively supercoiled (nSC)4The seed region is located in the center of the crRNAtarget duplex, where it is more sensitive to mismatches than the nonseed region (Abudayyeh et al, 16) Single mismatch can be fully tolerated by C2c2, but if double mismatches are located in the seed region, C2c2 is unable to cleave the singlestrand RNA;Using a mutagenesis assay in vivo, we discovered that a seed motif located at the tagdistal region of the crRNA is required by Cmr1α for target RNA capture by the Cmrα system thereby enhancing target specificity and efficiency These findings further refine the model for immune defence of Type IIIB CRISPRCas system, commencing on capture, cleavage and regulation



Figure 3 From Extensive Crispr Rna Modification Reveals Chemical Compatibility And Structure Activity Relationships For Cas9 Biochemical Activity Semantic Scholar
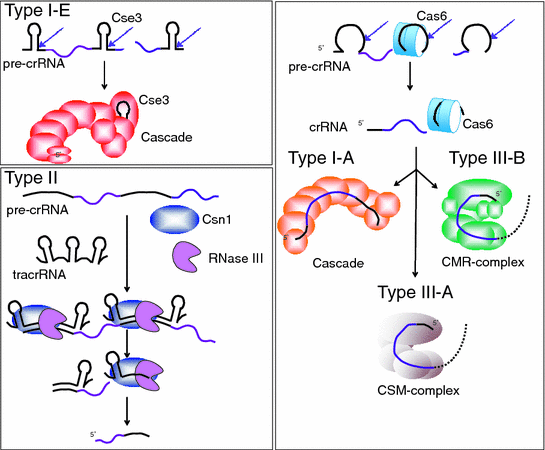


Crrna Biogenesis Springerlink
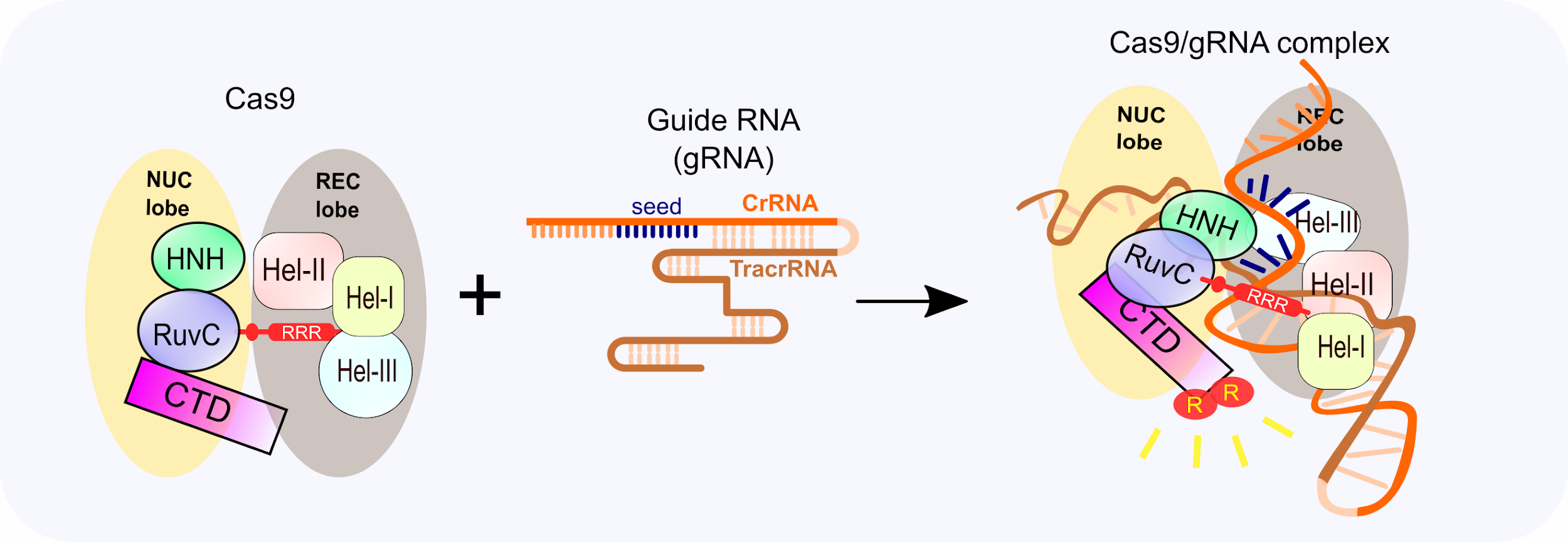
The guide RNA is a specific RNA sequence that recognizes the target DNA region of interest and directs the Cas nuclease there for editing The gRNA is made up of two parts crispr RNA (crRNA), a 17 nucleotide sequence complementary to the target DNA, and a tracr RNA, which serves as a binding scaffold for the Cas nucleaseMutations in the seed region abolish CRISPR/Cas mediated immunity by reducing the binding affinity of the crRNAguided Cascade complex to protospacer DNA We propose that the crRNA seed sequence plays a role in the initial scanning of invader DNA for a match, before base pairing of the fulllength spacer occurs, which may enhance the protospacer locating efficiency of the E coli Cascade complexDNA binding energetics is dominated by a subset of crRNA positions known as the "seed," which in E coli lies within the 5′terminal eight spacer nucleotides Mismatches in the seed are far more
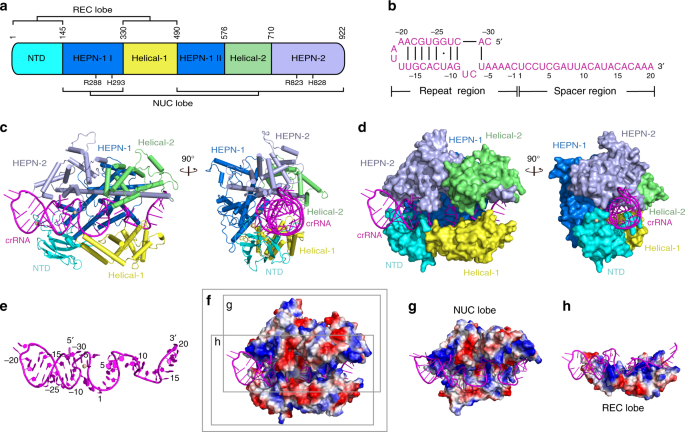


Two Hepn Domains Dictate Crispr Rna Maturation And Target Cleavage In Cas13d Nature Communications



Planting The Seed Target Recognition Of Short Guide Rnas Trends In Microbiology
Mutations in the seed region abolish CRISPR/Cas mediated immunity by reducing the binding affinity of the crRNAguided Cascade complex to protospacer DNA We propose that the crRNA seed sequence plays a role in the initial scanning of invader DNA for a match, before base pairing of the fulllength spacer occurs, which may enhance the protospacer locating efficiency of the E coli Cascade complexFour nucleotides in the flap region (red) of the aptamer form a complementary duplex with the crRNA seed sequence (blue) Consequently, Cas9's target DNA binding and complementary base pairing between target DNA and crRNA are inhibited, and the switch is turned offCRISPR regions are composed of multiple repeated sequences ranging from 21 to 48 bp in length separated by 26 to 72bp spacers (1, 2) The sequences of spacer regions are variable but are often identical to sequences found within phages, plasmids, or other foreign DNA



Structural Basis For Guide Rna Processing And Seed Dependent Dna Targeting By Crispr Cas12a Sciencedirect



Addgene Crispr Guide
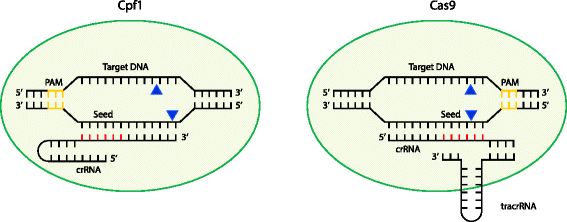
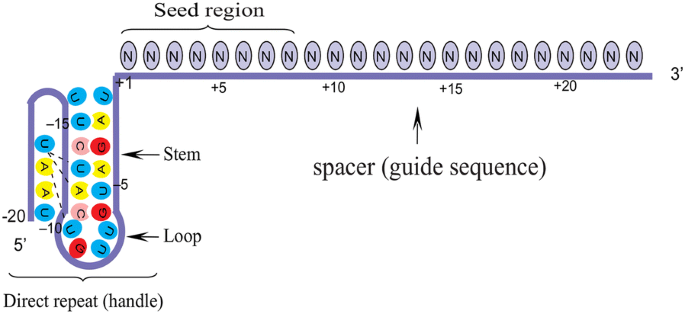
The minimal length of the basepairing region within the crRNA is 12 bp ("seed region"), which can in some cases lead to significant binding to other regions of the genome A recent study suggests that CRISPRbased genome editing has more offtarget effects than other genome editing tools such as TALENs or ZFNs (Fu et al, 13)Sequence recognition and structure of synthetic crRNA and tracrRNA A The target recognition part of the crRNA consists of nt, whereas the seed region is the 10 nt initiating the DNA binding nucleation B The tracrRNA consists of 67 nt comprising an antirepeat region complementary to the crRNArepeat and stem loops responsible for Cas9 bindingSome regions of the crRNA molecule are more important than others Nucleotides 15 and 78 at the 5' end of the spacer portion of the crRNa are known as the seed sequence The 5' end of the crRNA appears to be most important for binding foreign DNA targets



Systematic Analysis Of Crispr Cas9 Mismatch Tolerance Reveals Low Levels Of Off Target Activity Sciencedirect



Planting The Seed Target Recognition Of Short Guide Rnas Sciencedirect
Mutations in the seed region abolish CRISPR/Cas mediated immunity by reducing the binding affinity of the crRNAguided Cascade complex to protospacer DNA We propose that the crRNA seed sequence plays a role in the initial scanning of invader DNA for a match, before base pairing of the fulllength spacer occurs, which may enhance theHow the "seed" region within the guide RNA specifies DNA binding has remained unknown To determine how Cas9 assembles with and positions the guide RNA prior to substrate recognition, we solved the crystal structure of catalytically active Streptococcus pyogenes Cas9 (SpyCas9) in complex with an 85nt sgRNA at 29 Å resolution ( Fig 1 andThe seed region (nucleotides 1–8) is dispensable for RuvC activation, but the duplex of the central spacer (nucleotides 9–15) is required We captured the catalytic state of Cas12i2, with both


A Streamlined Crispr Cas9 Approach For Fast Genome Editing In Toxoplasma Gondii And Besnoitia Besnoiti Hehl Journal Of Biological Methods



Synthetic Crispr Rna Cas9 Guided Genome Editing In Human Cells Pnas
Propose that the seed region of crRNA guides prefer an Aformlike architecture whereas the tracrRNApairing (tracrpairing)repeatregioniscompatiblewithabroader arrayofmodifications,includingDNA,2FANnd2,5RNA Despite strong Aform conformational preference, relatively bulky modifications such as C2orC4OMe(iv) Introducing a mismatch into Cas13acrR can improve the fidelity of Cas13a;Although the target sequence of the cr3′pksP protospacer contains an additional adenine in the ΔakuB strain, this additional adenine is located outside the last 6 bp of the protospacer, known as the seed region



High Throughput Characterization Of Cascade Type I E Crispr Guide Efficacy Reveals Unexpected Pam Diversity And Target Sequence Preferences Genetics



The Biology Of Crispr Cas Backward And Forward Sciencedirect
A mismatchsensitive 'seed region' exists in the centre of the crRNA–target duplex in which consecutive mismatches impede cleavage of target RNA despite C2c2 tolerance to single mismatches across the spacer The C2c2 effector is composed of two HEPN domains with catalytic residues that cleave ssRNAs at varying distances outside the crRNA bindingThe 3′ end of the guide sequence, also known as the "seed region", plays a critical role in recognition of target sequence Thus, based on structural analysis, accessibility of the last three bases in the seed region was a prominent feature to differentiate functional sgRNAs from nonfunctional ones (Fig 1b) In addition, base accessibility in positions 51–53 was also significantly differentIn this study, overexpression of LwCas13a by lentivirus in glioma cells reveals that crRNA‐EGFP induces a "collateral effect" after knocking down the target gene in EGFP‐expressing cells EGFRvIII is a unique EGFR mutant subtype in glioma, and the CRISPR‐Cas13a system induces death in EGFRvIII‐overexpressing glioma cells



Rna Guide Complementarity Prevents Self Targeting In Type Vi Crispr Systems Sciencedirect



Systematic Analysis Of Crispr Cas9 Mismatch Tolerance Reveals Low Levels Of Off Target Activity Sciencedirect
CRISPR regions are composed of multiple repeated sequences ranging from 21 to 48 bp in length separated by 26 to 72bp spacers (1, 2) The sequences of spacer regions are variable but are often identical to sequences found within phages, plasmids, or other foreign DNAInterestingly, mismatches between the crRNA and DNA throughout the spacer length highly reduce cleavage activity of cCas12b in vitro, with an 18 nt PAMadjacent seed sequenceIn type II CRISPR systems, the seed region has been defined as the PAMproximal 10–12 nucleotides located in the 3′ end of the nt spacer sequence (15, 47, 48, 94) Mismatches in this seed region severely impair or completely abrogate target DNA binding and cleavage, whereas close homology in the seed region often leads to offtarget binding events even with many mismatches elsewhere ( 78 )



Crystal Structure Of A Crispr Rna Guided Surveillance Complex Bound To A Ssdna Target Science



Massively Parallel Crispri Assays Reveal Concealed Thermodynamic Determinants Of Dcas12a Binding Pnas
C2c2 can even tolerate three consecutive mismatches in the nonseed region ( Abudayyeh et al , 16 )Fect base pairing of only this part of crRNA (denoted "seed sequence") with the corresponding part of the protospacer (denoted "seed region") as well as an intact PAM sequence are required for interference by the E coli subtype CRISPR/ Cas system All other mutants were restricted on g8 CRISPRC2c2 can even tolerate three consecutive mismatches in the nonseed region ( Abudayyeh et al , 16 )
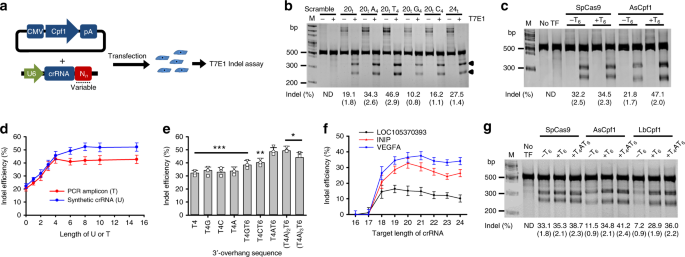


Highly Efficient Genome Editing By Crispr Cpf1 Using Crispr Rna With A Uridinylate Rich 3 Overhang Nature Communications
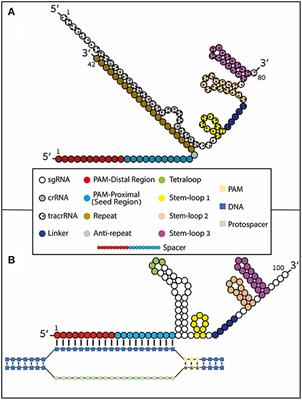


Frontiers Using Synthetically Engineered Guide Rnas To Enhance Crispr Genome Editing Systems In Mammalian Cells Genome Editing
Once the Cas9gRNA complex binds a putative DNA target, the seed sequence (810 bases at the 3′ end of the gRNA targeting sequence) will begin to anneal to the target DNA If the seed and target DNA sequences match, the gRNA will continue to anneal to the target DNA in a 3′ to 5′ directionCludes a ''seed'' region The seed region, which, for CRISPRCas enzymes, is a subset of nucleotides within the crRNA that base pairs with PAMproximal nucleotides, is highly sensitive against mismatches and is, therefore, critical for target affinity andspecificity(Gorskietal,17;Jineketal,12;Wiedenheft et al, 11)The seed region is located in the center of the crRNAtarget duplex, where it is more sensitive to mismatches than the nonseed region (Abudayyeh et al, 16) Single mismatch can be fully tolerated by C2c2, but if double mismatches are located in the seed region, C2c2 is unable to cleave the singlestrand RNA;
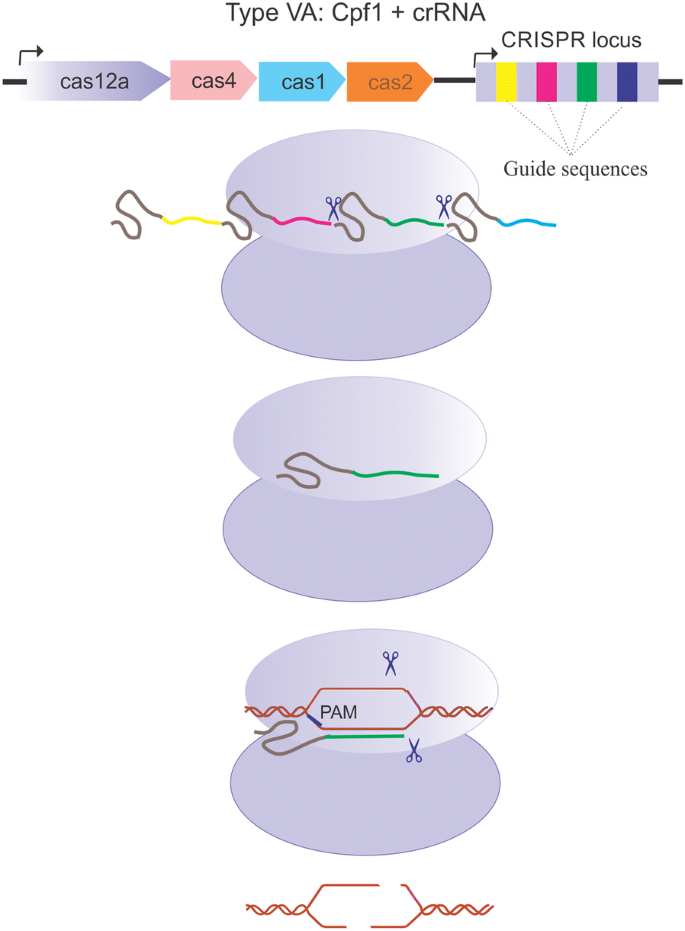


Crispr Cpf1 Proteins Structure Function And Implications For Genome Editing Cell Bioscience Full Text



The Influence Of Mismatches Between The Crrna And Target Dna On Binding Download Scientific Diagram
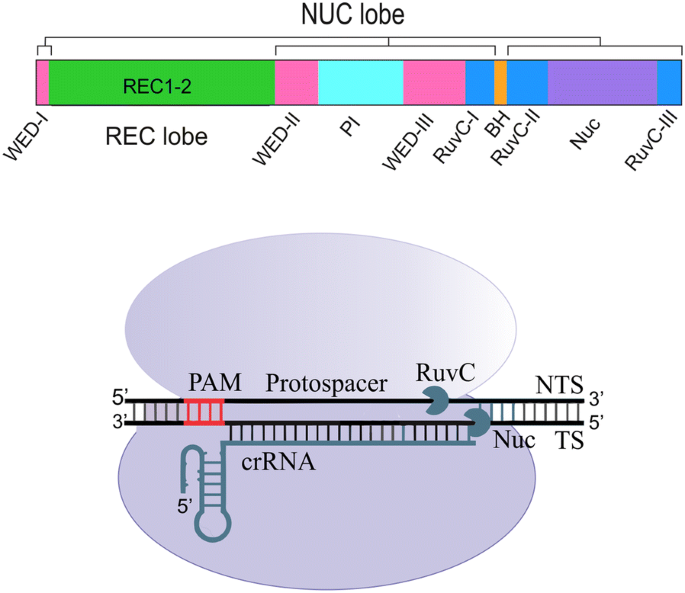
Cludes a ''seed'' region The seed region, which, for CRISPRCas enzymes, is a subset of nucleotides within the crRNA that base pairs with PAMproximal nucleotides, is highly sensitive against mismatches and is, therefore, critical for target affinity andspecificity(Gorskietal,17;Jineketal,12;Wiedenheft et al, 11)Seed segment binding and RNA–DNA duplex hybridization The location of seed segment of Cpf1 with 5–6 nt length is at the 5′ end of the crRNA spacer The seed segment is critical for the initial steps of sitespecific crRNAtarget strand hybridization Upon PAM recognition, local unwinding of the target DNA locates the PAMproximal target strand nucleotides in attachment with the seed segment(A) Schematic of central seed region of the guide in the crRNA binding the protospacer (target) region in the RNA (B) (Left) Schematic illustrating gRNAs targeting the SS and DS regions in the RNA



Overview Of The Crispr Cas9 Mechanism Of Action A Crispr Cas Download Scientific Diagram



Massively Parallel Crispri Assays Reveal Concealed Thermodynamic Determinants Of Dcas12a Binding Biorxiv
The seed region (nucleotides 2–8 from the 5′end) of the siRNA guide strand is known to downregulate the expression of genes outside of the canonical targets with weak interaction due to the seed sequence, which is commonly called 'offtarget' genes Thus, the mechanism of the offtarget effect due to siRNA is considered to be veryIn and around the crRNA 'seed' region were tolerated best A wider range of modifications were acceptable outside of the seed, especially D2 deoxyribose, and we exploited this property to facilitate exploration of greater chemical diversity within the seed 2 fluoro was the most compatible modThe crRNA 'seed' region, which is important for the Cas9mediated DNA recognition, is extensively recognized by the conserved arginine cluster in the bridge helix between the two lobes 50 ••



Base Complementarity Of The Pam Distal Target Region And The 5 Crrna Download Scientific Diagram
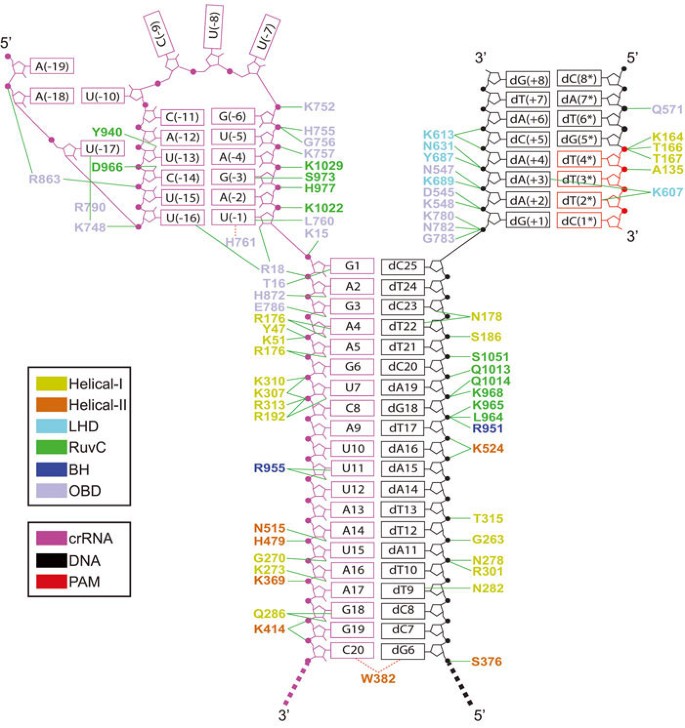


Type V Crispr Cas Cpf1 Endonuclease Employs A Unique Mechanism For Crrna Mediated Target Dna Recognition Cell Research
Spacers/protospacers In a bacterial genome, CRISPR loci contain "spacers" (viral DNA inserted into a CRISPR locus) that in type II adaptive immune systems were created from invading viral or plasmid DNA (called "protospacers") Upon subsequent invasion, a CRISPRassociated nuclease such as Cas9 attaches to a tracrRNA–crRNA complex, which guides Cas9 to the invading protospacer sequenceThe researchers reported highresolution structures of Cas12i2crRNA in three states, including the binding state, the seed region pairing state and the catalytic state These data suggested that



The Crispr Cas Revolution Reaches The Rna World Cas13 A New Swiss Army Knife For Plant Biologists Wolter 18 The Plant Journal Wiley Online Library


The Biology Of Native And Adapted Crispr Cas Systems Journal Of Young Investigators



Crispr Cas12a Functional Overview And Applications Sciencedirect


Crrna Processing Pathways In Type Ii Crispr Cas Systems In Type Ii Download Scientific Diagram



A Seed Interaction Is Required For Effective Interference To Download Scientific Diagram


Crispr Rt



Harnessing Crispr Cas Systems For Bacterial Genome Editing Trends In Microbiology



Scope Flexible Targeting And Stringent Carf Activation Enables Type Iii Crispr Cas Diagnostics Biorxiv



Partial Dna Replacement At The Guide Region Of A Gfp Crrna Induces Gene Download Scientific Diagram
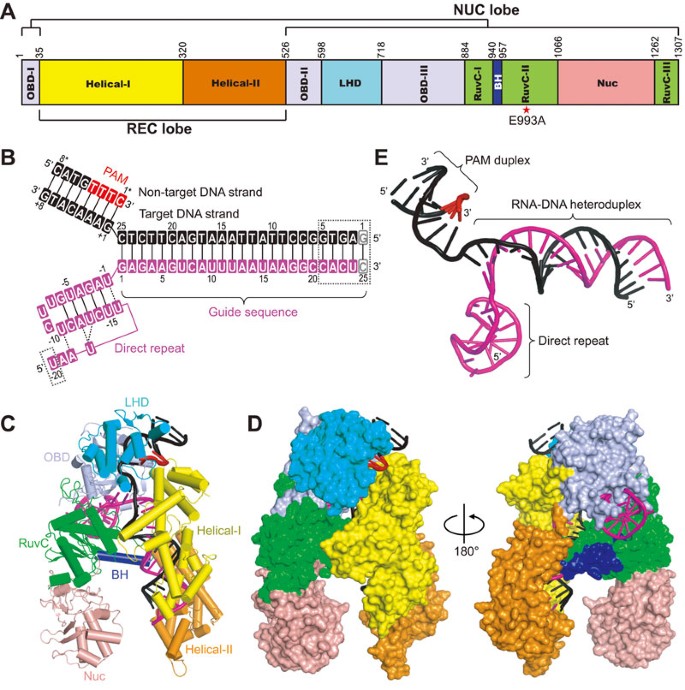


Type V Crispr Cas Cpf1 Endonuclease Employs A Unique Mechanism For Crrna Mediated Target Dna Recognition Cell Research



Interference By Clustered Regularly Interspaced Short Palindromic Repeat Crispr Rna Is Governed By A Seed Sequence Pnas


Chimeric Guides Probe And Enhance Cas9 Biochemical Activity Biochemistry X Mol



The Biology Of Crispr Cas Backward And Forward Sciencedirect



Figure 2 From Extensive Crispr Rna Modification Reveals Chemical Compatibility And Structure Activity Relationships For Cas9 Biochemical Activity Semantic Scholar


Team Tudelft Model Off Targeting 17 Igem Org



Rna Binding And Hepn Nuclease Activation Are Decoupled In Crispr Cas13a Sciencedirect



Crispr Cas12a Has Widespread Off Target And Dsdna Nicking Effects Journal Of Biological Chemistry


The Magic Cut On Target Dna By Crispr Cas9 Bioscope



High Throughput Characterization Of Cascade Type I E Crispr Guide Efficacy Reveals Unexpected Pam Diversity And Target Sequence Preferences Genetics


An Overview Of Designing And Selection Of Sgrnas For Precise Genome Editing By The Crispr Cas9 System In Plants Springerlink



Structures And Mechanisms Of Crispr Rna Guided Effector Nucleases Sciencedirect



Crispr Cas9 Guide Rna Specificity


Crispr Cpf1 Proteins Structure Function And Implications For Genome Editing Cell Bioscience Full Text



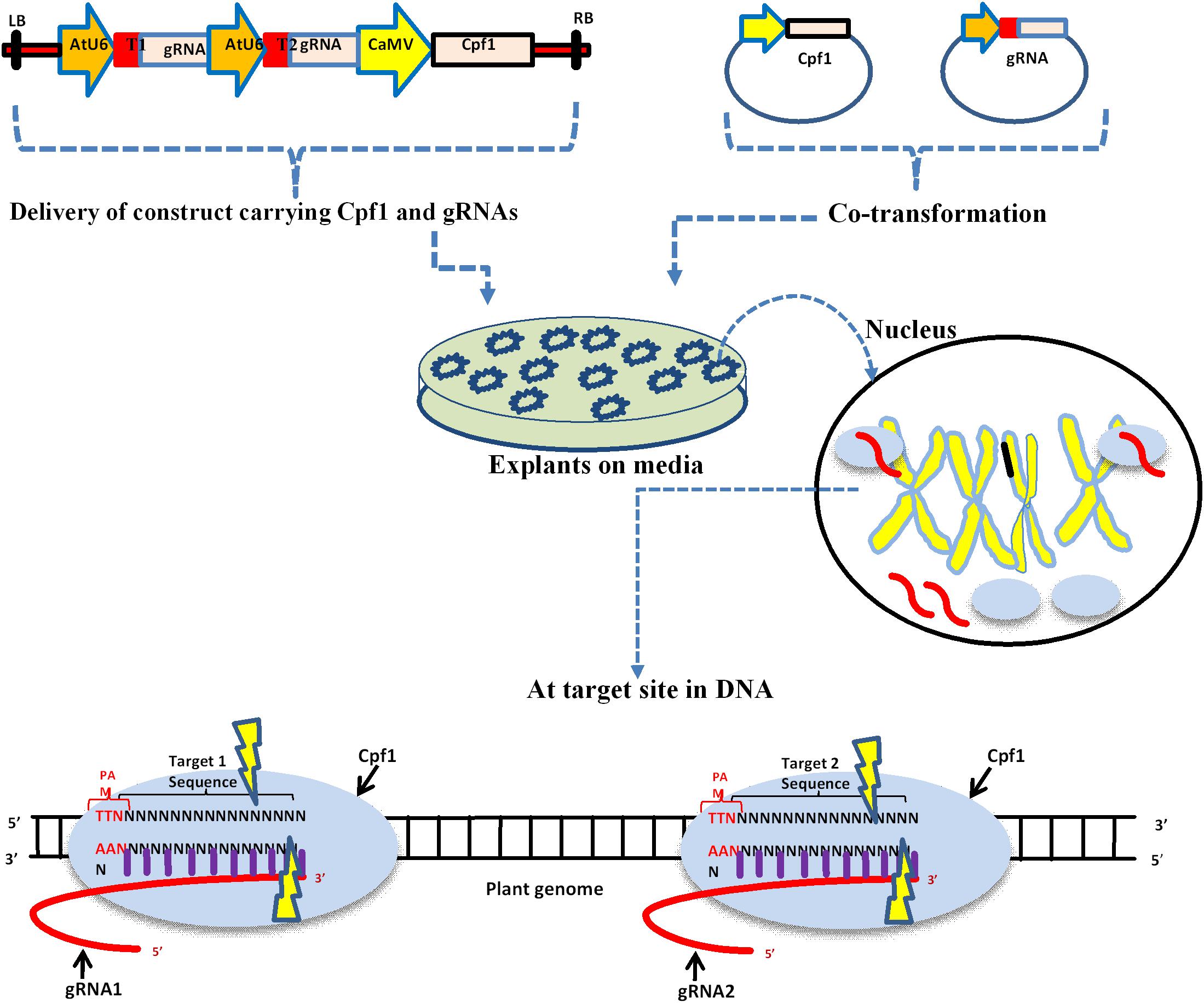
Design Principle Of A Crispr Cas9 Expression Vector For Construction Download Scientific Diagram



Streptococcus Pyogenes Crispr Spcas9 Guide Rna Anatomy A Endogenous Download Scientific Diagram


Frontiers Crispr Cas12a Cpf1 A Versatile Tool In The Plant Genome Editing Tool Box For Agricultural Advancement Plant Science
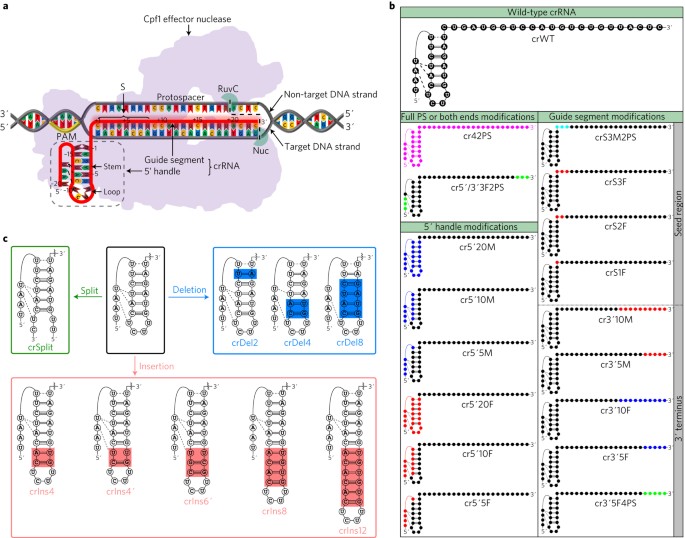


Engineering Crispr Cpf1 Crrnas And Mrnas To Maximize Genome Editing Efficiency Nature Biomedical Engineering
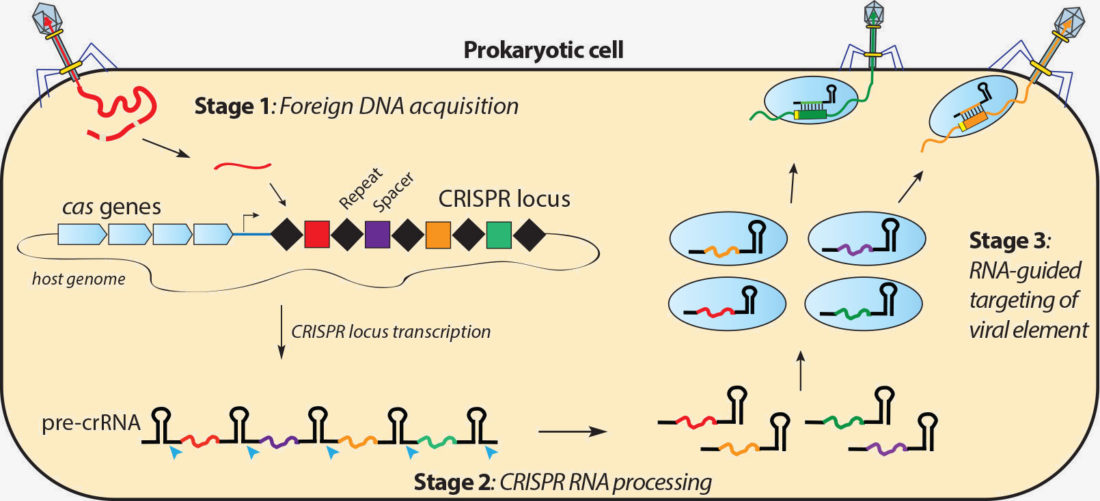


Crispr Systems Doudna Lab


Crispr Rt
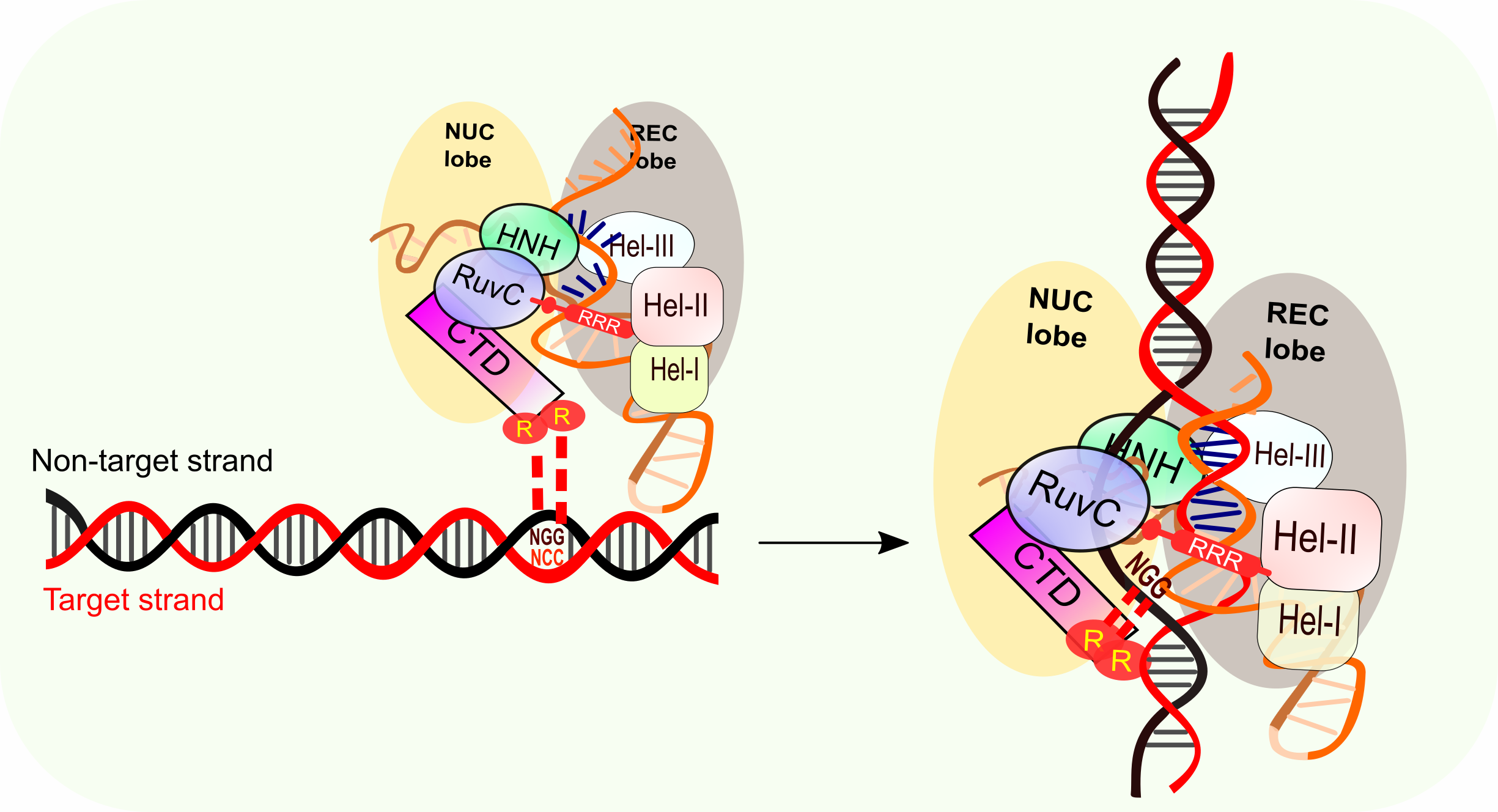


The Magic Cut On Target Dna By Crispr Cas9 Bioscope
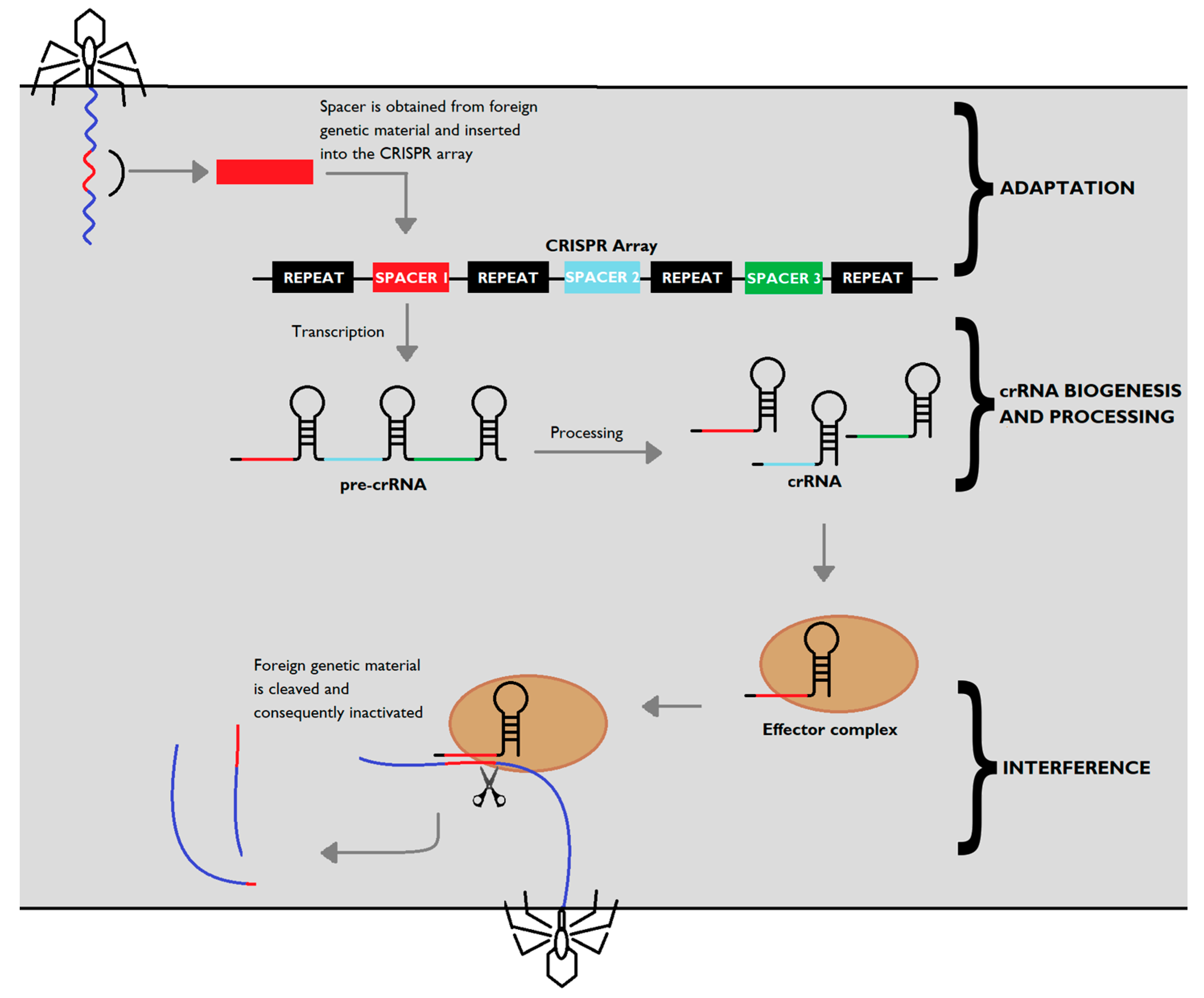


Antibiotics Free Full Text Crispr Cas Converting A Bacterial Defence Mechanism Into A State Of The Art Genetic Manipulation Tool Html



Degenerate Target Sites Mediate Rapid Primed Crispr Adaptation Pnas



Transforming Plant Biology And Breeding With Crispr Cas9 Cas12 And Cas13 Schindele 18 Febs Letters Wiley Online Library
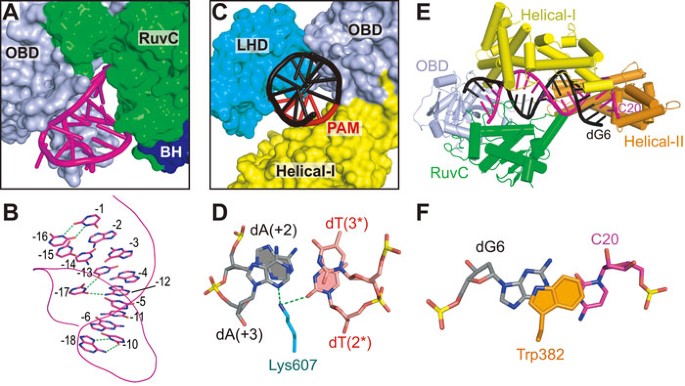


Type V Crispr Cas Cpf1 Endonuclease Employs A Unique Mechanism For Crrna Mediated Target Dna Recognition Cell Research



Kinetic Basis For Dna Target Specificity Of Crispr Cas12a Sciencedirect



Selection Of Dual Crrna Protospacer Sequences A Schematic Diagram Of Download Scientific Diagram



Massively Parallel Crispri Assays Reveal Concealed Thermodynamic Determinants Of Dcas12a Binding Biorxiv



Interference By Clustered Regularly Interspaced Short Palindromic Repeat Crispr Rna Is Governed By A Seed Sequence Pnas
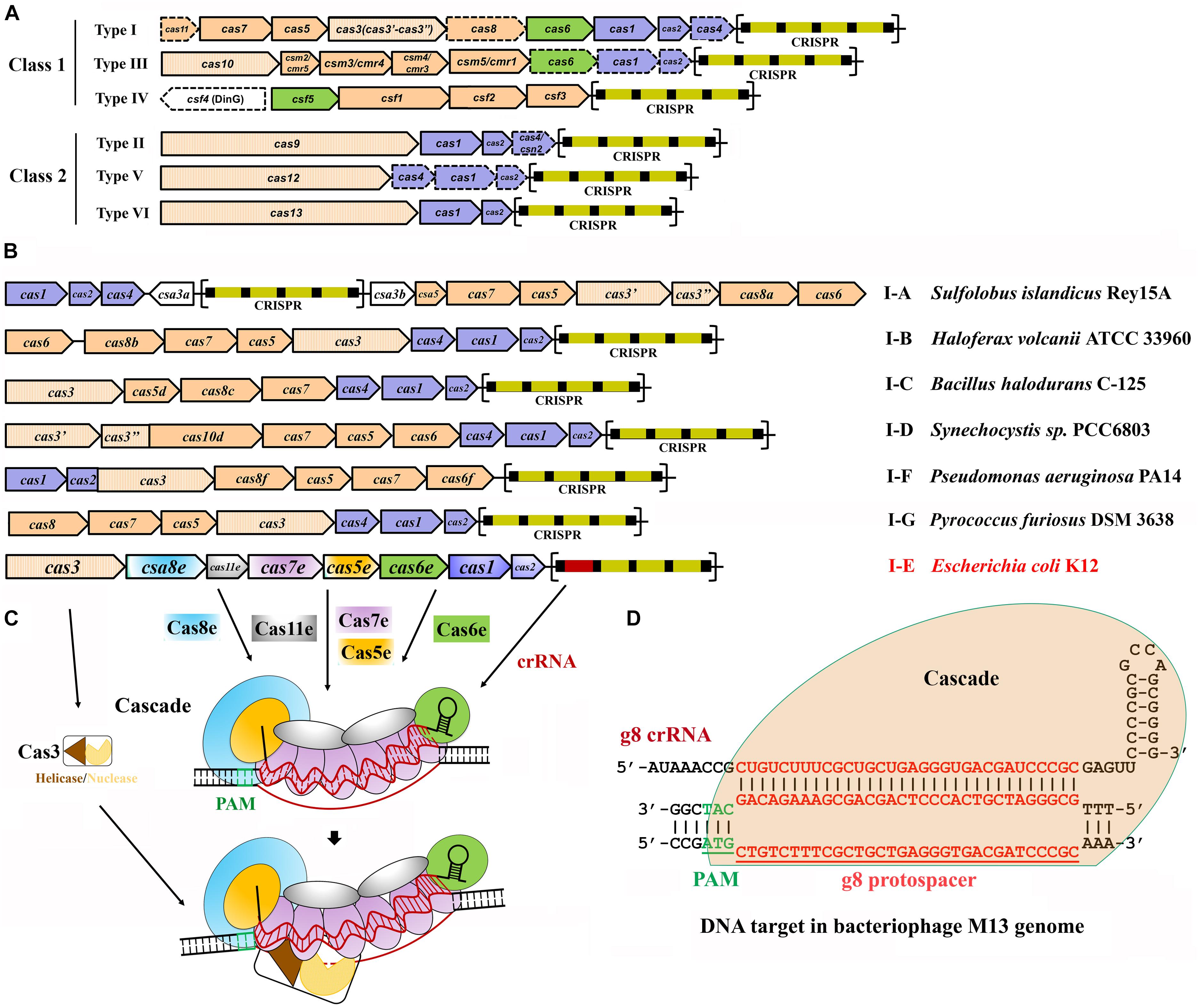


Frontiers Endogenous Type I Crispr Cas From Foreign Dna Defense To Prokaryotic Engineering Bioengineering And Biotechnology
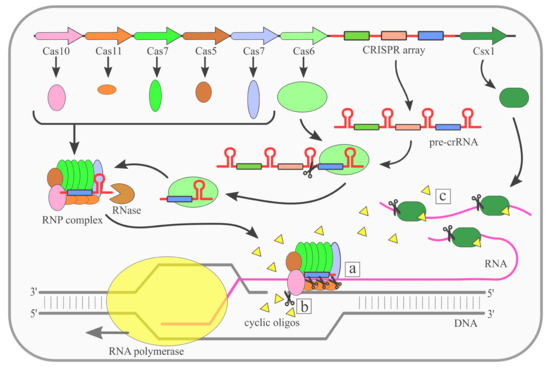


Ijms Free Full Text Rna Targeting Crispr Cas Systems And Their Applications Html


The Cpf1 Crispr Cas Protein Expands Genome Editing Tools Genome Biology Full Text


Rna Based Recognition And Targeting Sowing The Seeds Of Specificity Nature Reviews Molecular Cell Biology


Full Article Defining The Seed Sequence Of The Cas12b Crispr Cas Effector Complex



Partial Dna Replacement At The Guide Region Of A Gfp Crrna Induces Gene Download Scientific Diagram


Crispr Rt


Crispr Cpf1 Proteins Structure Function And Implications For Genome Editing Cell Bioscience Full Text



Scope Flexible Targeting And Stringent Carf Activation Enables Type Iii Crispr Cas Diagnostics Biorxiv



A Cas9 Guide Rna Complex Preorganized For Target Dna Recognition Science



Trans Activating Crrna An Overview Sciencedirect Topics



Binding Of Central Seed Region In Grna Crucial For Cas13 Mediated Download Scientific Diagram



The Biology Of Crispr Cas Backward And Forward Sciencedirect



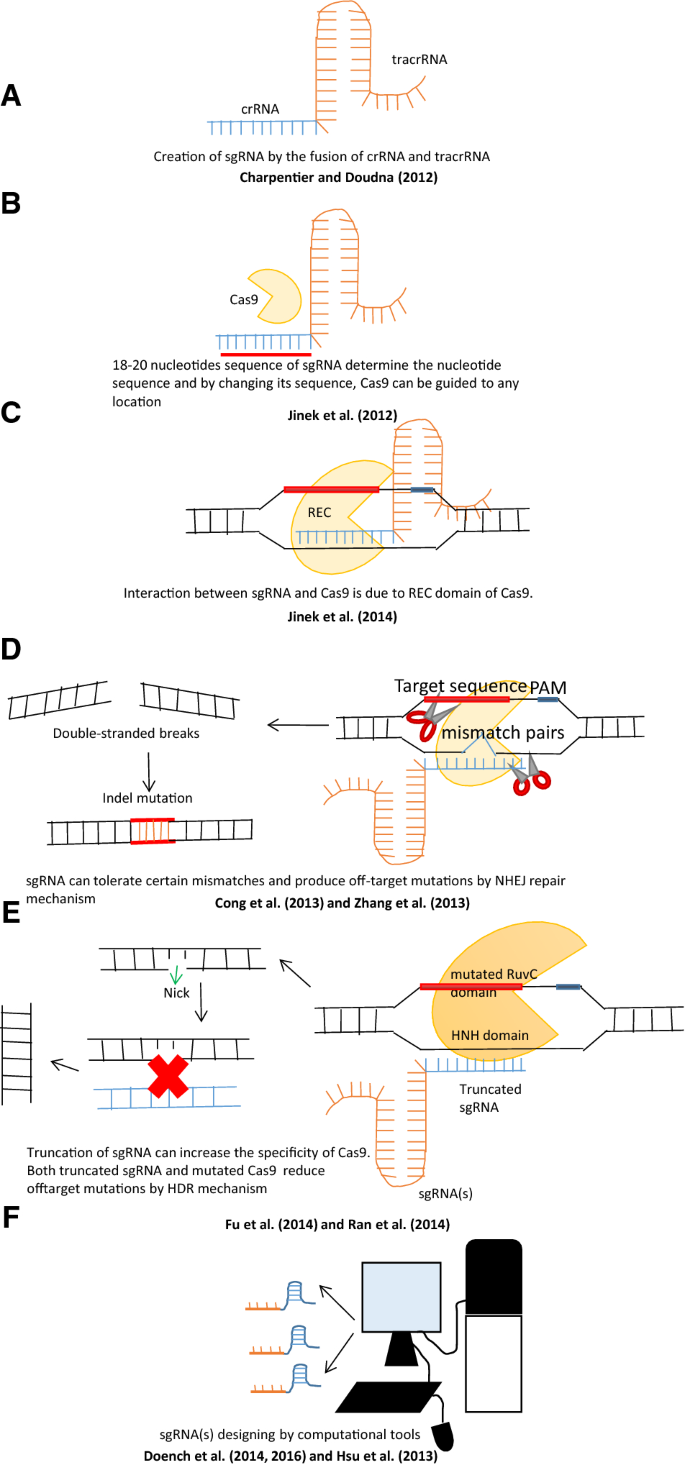
A Programmable Dual Rna Guided Dna Endonuclease In Adaptive Bacterial Immunity Science



The Revolution Continues Newly Discovered Systems Expand The Crispr Cas Toolkit Sciencedirect


Frontiers The Rise Of The Crispr Cpf1 System For Efficient Genome Editing In Plants Plant Science



Partial Dna Replacement At The Guide Region Of Crrna Or Sgrna Induces Download Scientific Diagram
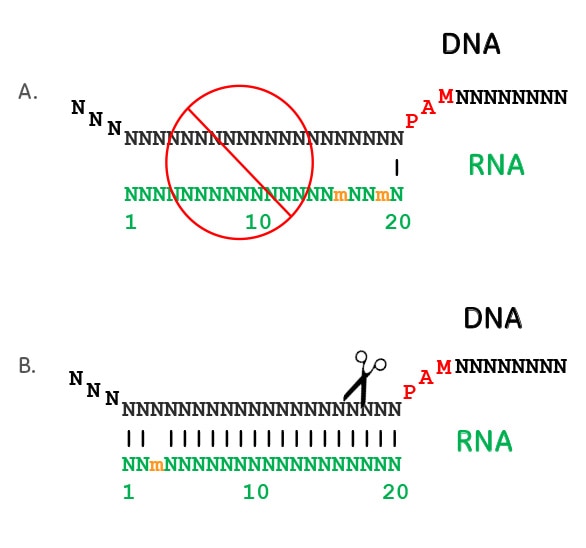


Crispr Cas9 Guide Rna Specificity
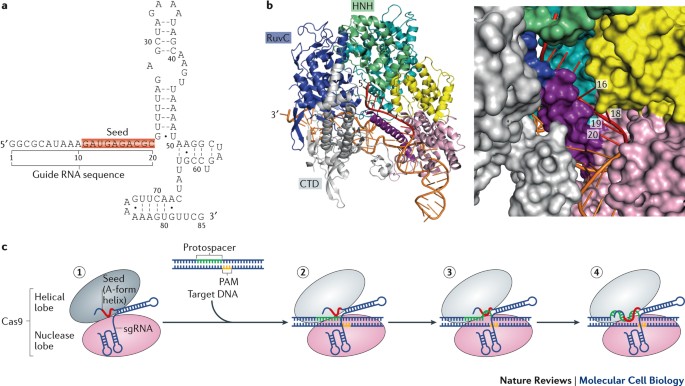


Rna Based Recognition And Targeting Sowing The Seeds Of Specificity Nature Reviews Molecular Cell Biology



Interference By Clustered Regularly Interspaced Short Palindromic Repeat Crispr Rna Is Governed By A Seed Sequence Pnas
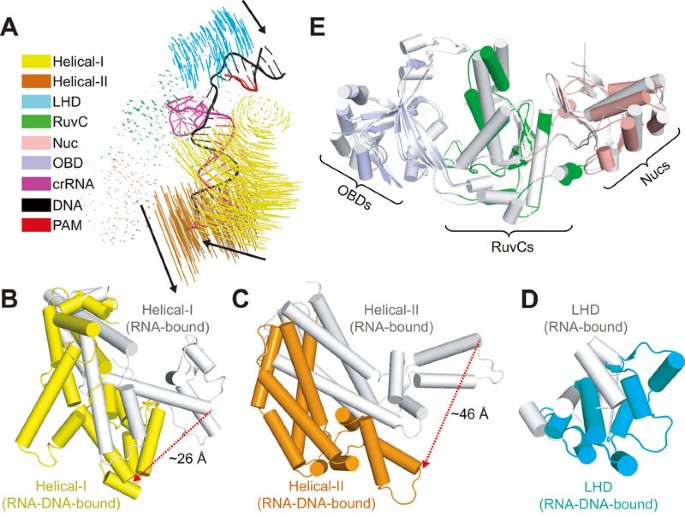


Type V Crispr Cas Cpf1 Endonuclease Employs A Unique Mechanism For Crrna Mediated Target Dna Recognition Cell Research
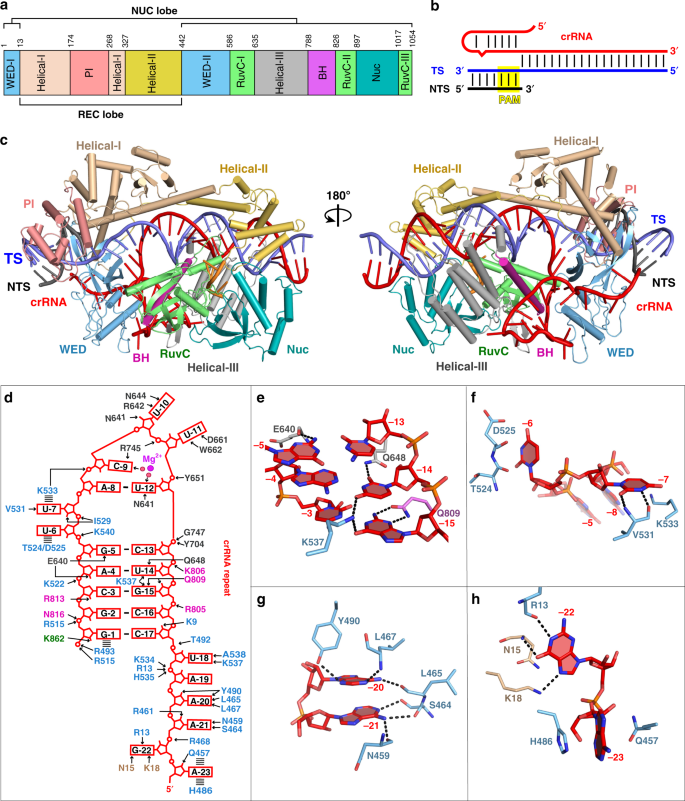


Structural Basis For Two Metal Ion Catalysis Of Dna Cleavage By Cas12i2 Nature Communications



Engineering The Direct Repeat Sequence Of Crrna For Optimization Of Fncpf1 Mediated Genome Editing In Human Cells Molecular Therapy



0 件のコメント:
コメントを投稿